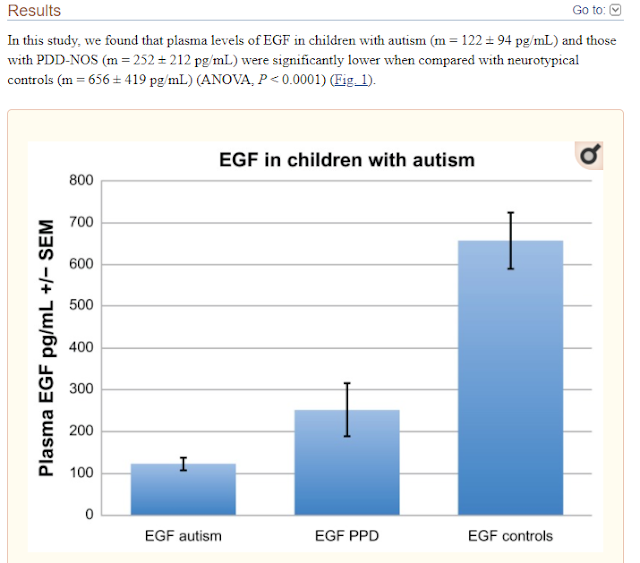
Today’s post combines a very simple therapy
for sound sensitivity that landed in my inbox from New Zealand with two more
genes that I was recently asked about.
Before I get started I would like to
thank our reader Daniel who is trying to spread that word that the IGF-1
targeting therapy cGPMax works in some Rett syndrome (half a capsule daily). I
did go into the science of IGF-1 related therapies at the recent conference in
Abu Dhabi. In that presentation I pointed out that the cGPMax therapy might
well be helpful in Pitt Hopkins syndrome. I saw today that Soko, an 8 year old
girl with Pitt Hopkins, had already made a trial and her parents are
impressed:-
“Equally
significant has been the positive shift in Soko's emotional well-being. Her
struggles with irritability and mood fluctuations feel like are not as frequent
and we feel like there is more often a sense of calm and emotional regulation.
This has had a profound ripple effect on our little family and our stress
levels.
Perhaps
most striking has been the accelerated rate at which Soko acquires new skills.
CGP Max has seemingly unlocked hidden potentials within her. This rapid skill
acquisition has been very exciting for us. In the last year she has gone from
being unable to walk to walking unassisted and even tackling steps no handed!”
I did some checking and some other
parents have tried cGPMax for Pitt Hopkins. For Rett syndrome Daniel found that
a lower dose was more beneficial than a higher dose. It is always best to start
with low doses and gradually increase them.
This does link to today’s post because
a microdeletion of NRXN1 can cause Pitt Hopkins Like Syndrome 2 (PHLS2).
In theory all these syndromes are untreatable, but try telling that to Soko’s
parents.
Back to sound
sensitivity
Today’s sound sensitivity is the type that
is moderated by Ponstan (mefenamic acid) and indeed Diclofenac. It might well
include those whose sound sensitivity responds to a simple potassium
supplement.
If you want to look into the details,
you can see from previous posts how potassium and potassium ion channels play a
fundamental role in both hearing and its sensory processing. They also play a
key role in excitability of neurons and so can play a key role in some epilepsy
and some intellectual disability.
It turns out that Cilantro/Coriander
leaves contains a chemical that activates the ion channels KCNQ2 (Kv7.2) and KCNQ3 (Kv7.3). This effect
is shared by Ponstan and Diclofenac.
In the case of Andy from New Zealand
the effect of a 425mg Cilantro supplement lasts very much longer than taking a
low dose of Ponstan or Diclofenac.
So, if your child responds well to
Ponstan and can then happily take off his/her ear defenders, but you do not
want to medicate every day, then a trial of Cilantro could be interesting.
I was curious as to why the effect
would last so much longer than Ponstan/Diclofenac. All of these drugs lower potassium levels
within neurons. Is the beneficial effect
coming from lowering potassium levels and so reducing neuronal
excitability? Or, is the effect coming
directly from a specific ion channel?
Nobody can tell you the half-life of
the active component of cilantro,
(E)-2-dodecenal, in humans. Andy
thinks it must have a long half-life.
Cilantro
(Coriander leaves)
If you live in North America you will
know what cilantro is, for everyone else it means coriander leaves. Coriander
seeds are the dried fruit of the coriander plant and, confusingly, in American
English coriander means coriander seeds.
The medicinal properties of the leaves
and seeds are not the same.
Cilantro leaves contain a compound
called (E)-2-dodecenal, which has been shown to activate a specific family of
potassium ion channel called KCNQ, otherwise known as Kv7 . These channels are
found in neurons, and they play an important role in regulating the electrical
activity of the brain.
When (E)-2-dodecenal binds to KCNQ/Kv7
channels, it causes them to open, which allows potassium ions to flow out of
the neuron. This outflow of potassium ions helps to stabilize the neuron's
membrane potential and makes it less likely to fire an action potential.
The level of potassium inside neurons
is much higher than the level outside. Having it too high, or indeed too low,
would affect the excitability of the neuron.
I am wondering if the problem with
potassium is mirroring the problem we have with chloride; perhaps both are
elevated inside neurons. That would be nice and simple.
The discovery that cilantro can
activate KCNQ channels helps to explain its potential anticonvulsant
properties. KCNQ channel dysfunction has
been linked to certain types of epilepsy, and drugs that activate these channels
are being investigated as potential treatments for these conditions.
Research suggests cilantro's
active compound, (E)-2-dodecenal, targets multiple KCNQ channels, particularly:
- KCNQ2/KCNQ3: This is the most
common type of KCNQ channel found in neurons.
- KCNQ1 in complex with
KCNE1: This
form is mainly present in the heart. KCNE1 acts as a regulatory subunit
that influences KCNQ1 channel function.
Cilantro leaf harbors a potent potassium channel-activating anticonvulsant
Herbs have a long history of use as folk medicine anticonvulsants, yet the underlying mechanisms often remain unknown. Neuronal voltage-gated potassium channel subfamily Q (KCNQ) dysfunction can cause severe epileptic encephalopathies that are resistant to modern anticonvulsants. Here we report that cilantro (Coriandrum sativum), a widely used culinary herb that also exhibits antiepileptic and other therapeutic activities, is a highly potent KCNQ channel activator. Screening of cilantro leaf metabolites revealed that one, the long-chain fatty aldehyde (E)-2-dodecenal, activates multiple KCNQs, including the predominant neuronal isoform, KCNQ2/KCNQ3 [half maximal effective concentration (EC50), 60 ± 20 nM], and the predominant cardiac isoform, KCNQ1 in complexes with the type I transmembrane ancillary subunit (KCNE1) (EC50, 260 ± 100 nM). (E)-2-dodecenal also recapitulated the anticonvulsant action of cilantro, delaying pentylene tetrazole-induced seizures. In silico docking and mutagenesis studies identified the (E)-2-dodecenal binding site, juxtaposed between residues on the KCNQ S5 transmembrane segment and S4-5 linker. The results provide a molecular basis for the therapeutic actions of cilantro and indicate that this ubiquitous culinary herb is surprisingly influential upon clinically important KCNQ channels
Activation of KCNQ5 by cilantro could also contribute to its gut stimulatory properties, as KCNQ5 is also expressed in gastrointestinal smooth muscle, and its activation might therefore relax muscle, potentially being therapeutic in gastric motility disorders such as diabetic gastroparesis.
The KCNQ activation profile of (E)-2-dodecenal bears both similarities and differences to that of other KCNQ openers. We recently found that mallotoxin, from the shrub Mallotus oppositifolius that is used in African folk medicine, also activates KCNQ1-5 homomers, prefers KCNQ2 over KCNQ3, and in docking simulations binds in a pose reminiscent to that predicted for (E)-2-dodecenal, between (KCNQ2 numbering) R213 and W236 In addition to the widespread use of cilantro in cooking and as an herbal medicine, (E)-2-dodecenal itself is in broad use as a food flavoring and to provide citrus notes to cosmetics, perfumes, soaps, detergents, shampoos, and candles (59).
Our mouse seizure studies suggest it
readily accesses the brain, and it is likely that its consumption as a food or
herbal medicine (in cilantro) or as an added food flavoring would result in
KCNQ-active levels in the human body;
we found the 1% cilantro extract an efficacious KCNQ activator, and
(E)-2-dodecenal itself showed greater than half-maximal opening effects on
KCNQ2/3 at 100 nM (.10 mV shift at this concentration) (EC50, 60 6 20 nM). We anticipate that its activity
on KCNQ channels contributes significantly to the broad therapeutic spectrum attributed
to cilantro, which has persisted as a folk medicine for thousands of years
throughout and perhaps predating human recorded history.
From the University of California:
How cilantro works as a secret weapon against seizures
In a new study, researchers uncovered the molecular action that enables cilantro to effectively delay certain seizures common in epilepsy and other diseases.
The
study, published in FASEB Journal, explains the molecular action of cilantro
(Coriandrum sativum) as a highly potent KCNQ channel activator. This new
understanding may lead to improvements in therapeutics and the development of
more efficacious drugs.
“We
discovered that cilantro, which has been used as a traditional anticonvulsant
medicine, activates a class of potassium channels in the brain to reduce
seizure activity,” said Geoff Abbott, Ph.D., professor of physiology and
biophysics at the UC Irvine School of Medicine and principal investigator on
the study.
“Specifically, we found one component
of cilantro, called dodecenal, binds to a specific part of the potassium
channels to open them, reducing cellular excitability.”
KCNQ channels and
autism
There is a growing body of
research suggesting a connection between KCNQ channels and autism.
·
KCNQ
channel mutations: Genetic
studies have identified mutations in several KCNQ channel genes (including
KCNQ2, KCNQ3) in individuals with ASD. These mutations might disrupt the normal
function of KCNQ channels, leading to abnormal brain activity.
- Neuronal excitability: KCNQ channels help regulate the electrical activity of neurons by controlling the flow of potassium ions. Mutations or dysfunction in KCNQ channels could lead to increased neuronal excitability, which has been implicated in ASD.
- Shared features: Epilepsy is a common
comorbidity with autism. Interestingly, KCNQ channel dysfunction is also
linked to certain types of epilepsy. This suggests some shared mechanisms
between these conditions.
KCNQ
Dysfunction and Intellectual Disability
Mutations in certain KCNQ
genes can lead to malfunctions in the corresponding potassium channels. These
malfunctions can disrupt normal neuronal activity and contribute to
intellectual disability.
- KCNQ2/3 Mutations: Research suggests increased activity in KCNQ2 and KCNQ3 channels, due to mutations in their genes, might be associated with a subset of patients with intellectual disability alongside autism spectrum disorder.
- KCNQ5 Mutations: Studies have
identified mutations in the KCNQ5 gene, leading to both loss-of-function
and gain-of-function effects on the channel. These changes in KCNQ5
channel activity can contribute to intellectual disability, sometimes
accompanied by epilepsy.
The other naming
system
KCNQ channels belong to a larger
potassium channel family called Kv7. So, you might see them referred to as
Kv7.1 (KCNQ1), Kv7.2 (KCNQ2), and so on, based on their specific gene and
protein sequence.
Mefenamic acid
and Kir channels (inwards rectifying potassium ion channels)
Ponstan (mefenamic acid) affects Kir
channels and KCNQ channels.
Different Kir channel subtypes contribute to
various brain functions, including:
- Neuronal excitability: Kir channels help
regulate the resting membrane potential of neurons, influencing their
firing activity.
- Potassium homeostasis: They play a role in
maintaining the proper balance of potassium ions within and outside
neurons, crucial for normal electrical signaling.
- Synaptic inhibition: Some Kir channels
contribute to inhibitory neurotransmission, which helps balance excitatory
signals in the brain.
Kir Channels are primarily inward rectifiers, meaning they allow potassium ions to flow more easily into the cell than out. They play a role in setting the resting membrane potential of cells, influencing their excitability.
KCNQ Channels can be voltage-gated or regulated by other factors. They contribute to various functions like regulating neuronal firing in the brain,
Other effects of
Cilantro
It is certainly could be just a
coincidence that Cilantro and Ponstan affect KCNQ channels. Cilantro has many
other effects.
Coriandrum sativum and Its Utility in Psychiatric Disorders
Recent research has shown that Coriandrum
sativum offers a rich source of metabolites, mainly terpenes
and flavonoids, as useful agents against central nervous system disorders, with
remarkable in vitro and in vivo activities on models related to these
pathologies. Furthermore, studies have revealed that some compounds exhibit a
chemical interaction with γ-aminobutyric acid, 5-hydroxytryptamine, and
N-methyl-D-aspartate receptors, which are key components in the pathophysiology
associated with psychiatric and neurological diseases.
Bioactivities of isolated compounds from Coriandrum
sativum by interaction with some neurotransmission systems
involved in psychiatric and neurological disorders.
Understanding
2p16.3 (NRXN1) deletions
One parent contacted me to ask about the genetic test results they had received for their child.
To understand what happens when parts
of the NRXN1 gene are missing you need to read up on neurexins and neuroligins.
Neurexins and
Neuroligins
Neurexins ensure the formation of
proper synaptic connections, fine-tune their strength, and contribute to the
brain's adaptability. Understanding their role is crucial for understanding
brain development, function, and various neurological disorders.
Neurexins and neuroligins are cell
adhesion molecules that work together to ensure proper synapse formation,
function, and ultimately, a healthy and functioning brain.
Neuroligins are located on the
postsynaptic membrane (receiving neuron) of a synapse.
Neurexins are located on the
presynaptic membrane (sending neuron) of a synapse.
Mutations in either neurexin or
neuroligin genes have been linked to various neurodevelopmental disorders,
including autism.
A comprehensive presentation for
families is below:
Understanding
2p16.3 (NRXN1) deletions
A microdeletion in the NRXN1
gene on chromosome 2p16.3 can cause a condition similar to Pitt-Hopkins
syndrome, but referred to as Pitt-Hopkins like syndrome 2 (PHLS2).
NRXN1 Gene:
- NRXN1 codes for a
protein called neurexin 1 alpha, which plays a critical role in the
development and function of synapses, the junctions between neurons in the
brain.
- Neurexin 1 alpha helps
neurons connect with each other and transmit signals.
Microdeletion:
- A microdeletion is a
small deletion of genetic material from a chromosome.
- In PHLS2, a
microdeletion occurs in the NRXN1 gene, removing some of the genetic
instructions needed to produce functional neurexin 1 alpha protein.
Pitt-Hopkins
Like Syndrome 2 (PHLS2):
- PHLS2 is a genetic
disorder characterized by intellectual disability, developmental delays,
and various neurodevelopmental features.
- Symptoms can vary
depending on the size and specific location of the NRXN1 microdeletion.
- Common features
include:
- Intellectual
disability (ranging from mild to severe)
- Speech and language
impairments
- Developmental delays
in motor skills
- Stereotypies
(repetitive movements)
- Seizures
- Behavioral problems
(e.g., hyperactivity, anxiety)
- Distinctive facial
features (not always present)
What has this got
to do with Pitt Hopkins syndrome (loss of TCF4)?
“TCF4
may be transcribed into at least 18 different isoforms with varying N-termini,
which impact subcellular localization and function. Functional analyses and
mapping of missense variants reveal that different functional domains exist
within the TCF4 gene and can alter transcriptional activation of downstream
genes, including NRXN1 and CNTNAP2, which cause Pitt-Hopkins-like syndromes 1
and 2.”
NRXN1
interactions with other genes/proteins
Given the function of neurexins and
neuroligins, you would expect that the common interactions of NRXN1 are with
neuroligins. We see below the NLGNs (neuroligin genes/proteins)
Our more avid readers may recall that neuroligins are one mechanism for regulating the GABA switch. This is the developmental switch that should occur in all humans about two weeks after birth. If it does not occur, the brain cannot develop and function normally. Autism and intellectual disability are the visible symptoms.
An unexpected role of neuroligin-2 in regulating
KCC2 and GABA functional switch
We report here that KCC2 is unexpectedly regulated by neuroligin-2 (NL2), a cell adhesion molecule specifically localized at GABAergic synapses. The expression of NL2 precedes that of KCC2 in early postnatal development. Upon knockdown of NL2, the expression level of KCC2 is significantly decreased, and GABA functional switch is significantly delayed during early development. Overexpression of shRNA-proof NL2 rescues both KCC2 reduction and delayed GABA functional switch induced by NL2 shRNAs. Moreover, NL2 appears to be required to maintain GABA inhibitory function even in mature neurons, because knockdown NL2 reverses GABA action to excitatory.
Our data suggest that in addition to its
conventional role as a cell adhesion molecule to regulate GABAergic
synaptogenesis, NL2 also regulates KCC2 to modulate GABA functional switch and
even glutamatergic synapses. Therefore, NL2 may serve as a master regulator in
balancing excitation and inhibition in the brain.
It would seem plausible that in the
case of microdeletions of the NRXN1 gene there will be a direct impact on the
expression of NLGN2 gene that encodes neuroligin 2.
So plausible therapies to trial for
microdeletions of the NRXN1 gene would include bumetanide, as well as cGPMax,
due to the link with Pitt Hopkins.
GPC5 gene
Finally, we move on to our last gene
which is GPC5.
The protein Glpycan 5/GPC5 plays a
role in the control of cell division and growth regulation.
Not surprising, GPC5 acts a tumor
suppressor, making it a cancer gene. Because of this it is also an autism gene.
It also plays a role in Alzheimer’s disease.
I was not sure I would be able to say
anything about how you might treat autism caused by a mutation in GPC5.
Glycan
susceptibility factors in autism spectrum disorders
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5556687/
I am assuming the mutation causes a
loss of function, meaning there is a reduced level of the protein Glpycan 5.
Since one role of this gene is to
suppress Wnt/beta-catenin signaling, you might want to replace this action.
This is actually covered in my blog in
various places. One way is via a GSK-3β inhibitor.
GSK-3β inhibitor include drugs
designed to block GSK-3β activity, examples include lithium (used for bipolar
disorder), kenpaullone, and tideglusib. Certain natural compounds like curcumin
and quercetin have been shown to possess GSK-3β inhibitory effects.
Atorvastatin, which my son has taken
for 10 years, is indirectly a GSK-3β inhibitor
Some natural compounds like fisetin
(found in fruits and vegetables) have been shown to promote beta-catenin
phosphorylation, leading to its degradation.
In previous posts I pointed out that
the cheap kids’ anthelmintic medication Mebendazole is indirectly another Wnt
inhibitor. This is because it reduces TNIK. TNIK promotes Wnt signaling by
stabilizing beta-catenin, a key player in the pathway. By reducing TNIK levels,
mebendazole indirectly disrupts Wnt signaling. Mebendazole is therefore a novel
cancer therapy and is being investigated to treat brain cancers, colon cancer,
breast cancers etc.
Unlike what is says in the literature
about GPC5, there actually are many
options that can be safely trialed.
Note that you may not know for sure
that any mutation is actually causal/pathogenic. Some people have several “likely
pathogenic” mutations, some likely are not.
Conclusion
We have covered the potassium ion
channel Kv7.1 previously. In Pitt Hopkins syndrome this ion channel is over
expressed and so you would want to inhibit it. Do not take Cilantro, it would
have the opposite effect to what you want.
It looks like cGPMAX is one thing you
need to trial for Pitt Hopkins syndrome and Rett syndrome. For idiopathic
autism it may, or may not help. Try a low dose first, observe the effect, then
try a higher dose.
In Rett syndrome we know that people
with have as much NKCC1 RNA — a molecule that carries the instructions to make
the protein — as healthy individuals. However, their levels of KCC2 RNA are
much lower, potentially disrupting the excitation/inhibition balance of nerve
cell signaling. This will result in elevated chloride in neurons. This is
correctable today using bumetanide.
People with NRXN1 microdeletions do
seem to have treatment options, as do people with GPC5 mutations.
Note that out reader Janu, treating a
mutation in GABRB2, reports success with a combination of the SSRI drug Lexapro
and sodium valproate.
I am a fan of low dose Ponstan for
sound sensitivity, it has numerous potentially beneficial mechanisms. It has
been even shown to protect against Alzheimer’s disease. There is no reason not to give cilantro a try
as an alternative or complement to improve sound sensitivity.
Dried coriander is normally made from
the seeds and is not what you need. In your supermarket you can buy fresh
coriander leaves (Cilantro). The fresh herb is about 90% water, but when you
dry the herb you will lose at lot of the active substance because it is
volatile and will evaporate. My guess is that you will need 2-3 g of the fresh
herb to equal Andy’s 425mg supplement. You
can eat the stalks as well as the leaves, it all has the same pungent taste.